Previously, we discussed the maximum a posteriori estimator and the maximum likelihood estimator. We gathered that using given data, we can make use of Bayes’ theorem and make a solid guess at parameters of a probability distribution, given the data we got and any prior knowledge. Even if we had no prior distribution, we could just get a solid guess thanks to MLE.
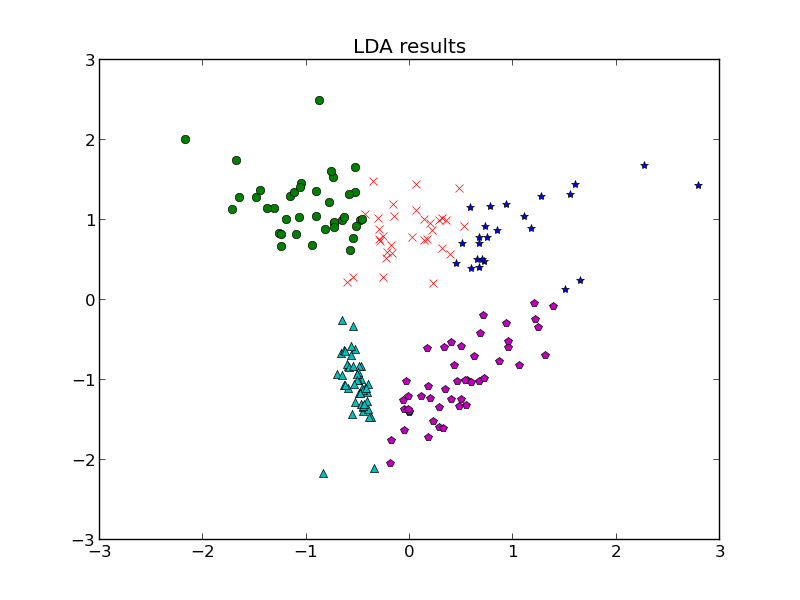
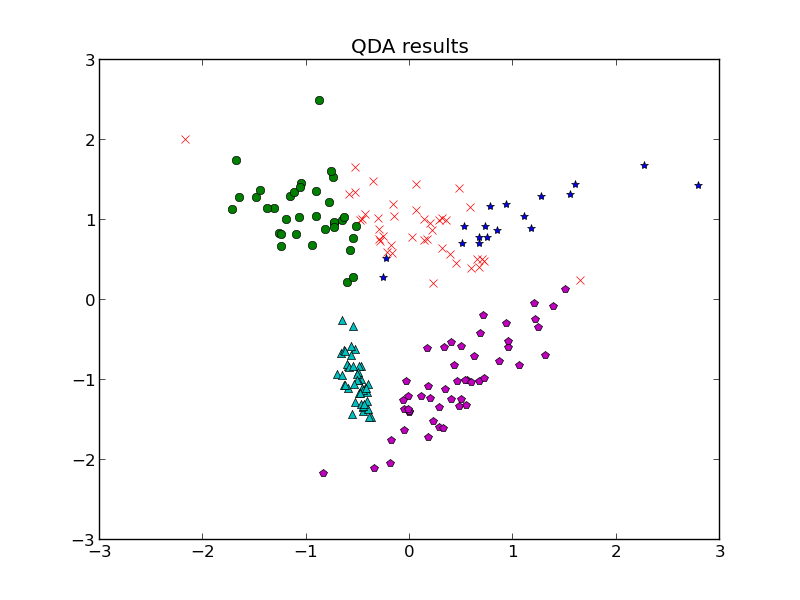
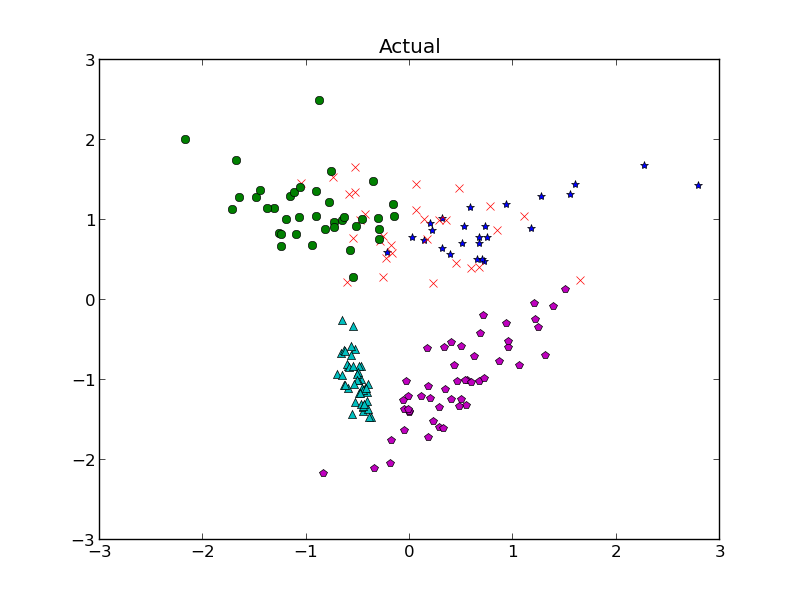
But let’s say the situation got a little more complicated. Last time, we discussed classification by linear discriminant analysis (LDA) and quadratic discriminant analysis (QDA), but those technically require us to have the parameters for the distributions of each class known ahead of time. So what if we need to guess that? Each class is going to have a distribution, and we have a bunch of data with no known classes, so how can we bridge the gap and not only classify our data but make a guess at the parameters of each class?
So the Expectation-Maximization algorithm gives us a method to basically try our best to get all the things we want to solve for above, landing at a possible local solution but maybe not the actual answer. Which is fine, we’re just making a guess. Sometimes that’s the best we can do.
Informally, what the algorithm entails is looping the E and M steps until convergence.
1. Expectation step: Using the prior from the previous step, find the expected value of the underlying variables we need in our setup; e.g. for classification, let’s classify the points by choosing the class we would expect each data point to be in, given our prior distributions from the previous step.
2. Maximization step: using the underlying variables we just solved for in the E step, use maximum likelihood estimators to find parameters for the distributions of whatever we’re looking for, e.g. for classification, the distribution of the classes.
In more formal math terms, lets say X is a vector of data, Z is the underlying variables we need, is the log likelihood function for parameters, given X and Z, and
is a vector of parameters we want to solve for. Breaking the algorithm up into 2 steps like before,
1. Expectation step; calculate the expected value
2. Maximization step; calculate new
So how would we do this for our little Gaussian mixture model we wanted to classify in the previous post? We have m classes, n data points (for now, let’s assume n >> m, we have tons of data). Making guesses at our parameters initially for each class, we do EM by repeating the following 2 steps.
1. Expectation step: find which class a point belongs to using LDA or QDA classifiers from the previous post (the assumptions each one assumes applied here, so choose one and stick with it).
2. Maximization step: for each class, use the points associated with it to find the sample mean. For LDA, we find the sample covariance matrix using all points so this won’t change between iterations, but for QDA, we need to also find the sample covariance matrix for each class.
If we wanted to entire ignore covariance in this calculation, you could actually see that k-means using a simple distance measure to decide on classes for the expectation step is basically an expectation maximization algorithm! Additionally, you can see that Baum-Welch is a variant of this for Hidden Markov Models where during our expectation step, we guess what states our system is in, and then use that to make a guess at our observation and state transition matrices.
Couple things: clearly, EM is a very general idea; the algorithm details need to be changed depending on the problem, i.e. things aren’t just Gaussian all the time, we need to adjust EM to suit our problems. EM algorithm tends to fall into a local solution but not necessarily the solution you want, and of course this changes when different priors and starting points are involved in the problem formulation. In the case of Baum-Welch and some other cases, EM algorithm isn’t even guaranteed to converge. So the EM algorithm needs to be used with care, though it is extremely powerful and has been proved to have some level of correctness.